
|
| Introduction | Interface Manual | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
E3Miner E3Miner is a text mining tool for ubiquitin-protein ligases (E3s). The tool is performed through the input of E3-related text. 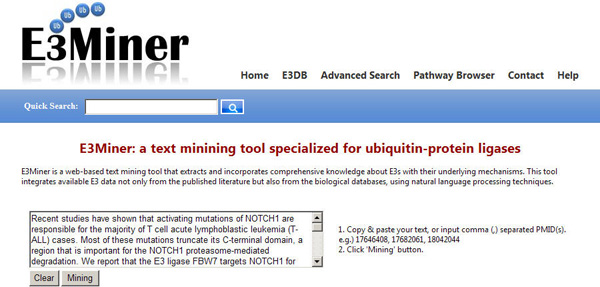
USAGE: You can insert text or comma (,) separated PubMed IDs (PMIDs),
and click the 'Mining' button. When a PMID is given, E3Miner makes online access to PubMed,
and retrieves the corresponding title and abstract.
Note that input text or PMIDs cannot begin with hyphen or comma (,).
E3 data mined is structured as a table form, and you can also save the E3 data in text format through
the 'Save Result' link. E3 data consist of the items as following.
('Heading' indicates the head in downloadable text data.)
EXAMPLE 1: Given the text of journal abstract
(PMID:17646408), E3Miner extracts E3 names and identifies their UniProt IDs with relevant data, as shown below.
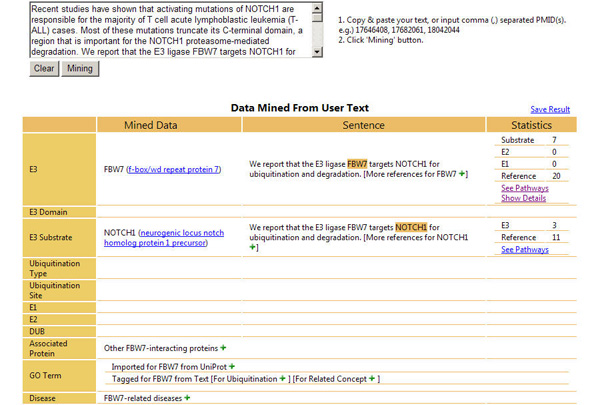
EXAMPLE 2: You can also see a video clip for an example usage of E3Miner,
which mines data about the TRAF6 gene from the text
(PMID:18042044),
and then examines TRAF6-related substrates, functions, and diseases from the output by E3Miner.
Quick Search Quick Search provides full-text search in all fields. This is always accessible through the search bar as shown below. 
USAGE: You can type any keywords with 'AND' and 'OR' operators.
If no operators are specified, then the keywords are treated as having the 'OR' operator.
When using 'AND' and 'OR' together, the operator 'AND' has a higher precedence than 'OR'.
Note that the keywords can be double-quoted for the exact match.
In the results, each icon represents links as follows. 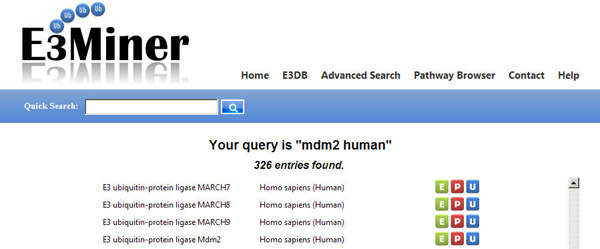
Advanced Search provides text search by a combination of selected fields. This is accessible by the menu tab 'Advanced Search'. 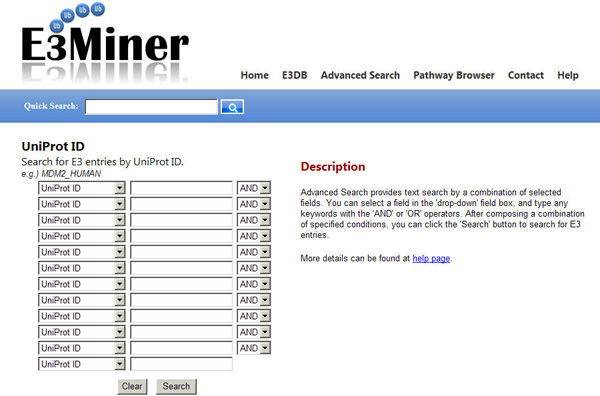
USAGE: You can select a field in the 'drop-down' field box, and type any keywords with
the 'AND' or 'OR' operators.
After composing a combination of specified conditions, you can click the 'Search'
button to search for E3 entries.
EXAMPLE: To search for "RING domain E3 protein that is related to human papilloma", you can input 'human' in the organism field, 'papilloma' in the disease field, and 'ring' in the E3 domain field. Then, you can get the results as shown below. 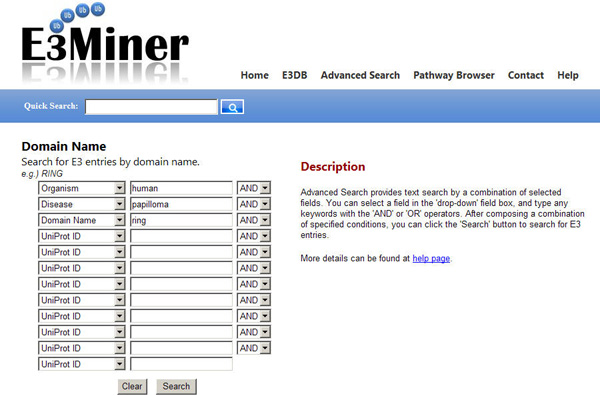
Pathway Browser Pathway Browser is a graphic tool to view the ubiquitination pathways of a particular E3.
USAGE: You can see the ubiquitination pathway from E1 to E2, E3, and the target protein.
You can control the distance of protein interactions in pathways by the "Zoom In"
|
| ©2007~2013 NLP*CL Laboratory, KAIST. All rights reserved. |
 : E3DB entry
: E3DB entry : Pathway Browser
: Pathway Browser : UniProt entry
: UniProt entry E1
E1 E2
E2 E3
E3 Target Protein
Target Protein