
|
| Introduction | Interface Manual |
|
|
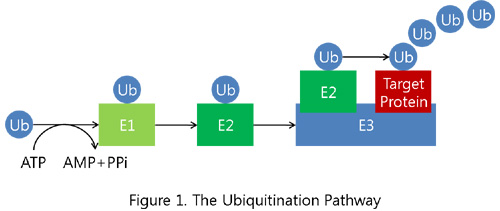
Ubiquitination, or ubiquitylation, is a common process of post-translational modification, which transfers ubiquitin into the target protein for a wide range of cellular effects in eukaryotes. As shown in Figure 1, ubiquitin is activated in an ATP-dependent manner by a ubiquitin-activating enzyme (E1), and it is transferred to a ubiquitin-conjugating enzyme (E2) that, with the help of a ubiquitin-protein ligase (E3), attaches ubiquitin to the target protein through the epsilon-amino group of a lysine residue. E3 is particularly responsible for the specific recognition of the target protein in a pathway. The attached ubiquitin chain is lengthened by the E2 and E3, sometimes with the help of an additional factor, i.e., the ubiquitin-chain elongation factor (E4).
* It is known that each ubiquitin-like modifier protein has a single dedicated E1. (Pickart and Eddins, 2004)[1] Glossary Ubiquitination pathway A pathway of ubiquitin to the target protein via enzyme cascades.
Ubiquitin: A small protein consisting of 76 amino acid residues, well conserved in eukaryotic cells. E3 domain E2-binding domains that catalyze ubiquitination.
The families of E3 proteins are generally classified by the E2-binding domains. (Ardley and Robinson, 2005)[2]
Ubiquitin structure Distinct forms of ubiquitins that are attached to the target protein.
Monoubiquitination: addition of a ubiquitin to a single lysine residue in the target protein. * There is no agreement on the use of these terms. For example, in some articles, 'multiubiquitination' is also used as 'polyubiquitination' with a similar meaning. Similarly, 'oligoubiquitination' is used with 'monoubiquitination'. We follow the terms used in (Welchman et al, 2005)[4] Human disease E3-dependent human diseases. E3s are known either to induce diseases directly, or to cause them indirectly by interacting with disease-induced proteins. Such diseases include cancers, genetic disorders, and viral infections. References [1] Pickart C.M. and Eddins M.J. (2004). Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55-72. (PMID: 15571809) [2] Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15-30. (PMID: 16250895) [3] Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003 Aug;21(8):921-6. (PMID: 12872131) [4] Welchman RL, Gordon C, Mayer RJ., Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005 Aug;6(8):599-609. Review. (PMID: 16064136) |
| ©2007~2013 NLP*CL Laboratory, KAIST. All rights reserved. |